| Name: | Chicken polyclonal antibody to fibrillarin |
| Immunogen: | Full length human fibrillarin expressed in and purified from E. coli. |
| HGNC Name: | FBL |
| UniProt: | P22087 |
| Molecular Weight: | 34.5kDa |
| Host: | Chicken |
| Isotype: | |
| Species Cross-Reactivity: | Human, Rat, Mouse, Dog, Horse |
| RRID: | AB_2572216 |
| Format: | Concentrated IgY preparation in PBS plus 0.02% NaN3 |
| Applications: | WB, IF/ICC, IHC |
| Recommended Dilutions: | WB: 1:2,000-1:5,000. IF/ICC: 1:2,000-1:5,000. IHC not recommended |
| Storage: | Store at 4°C |
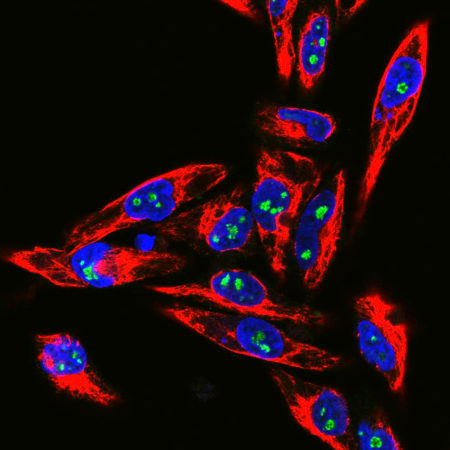
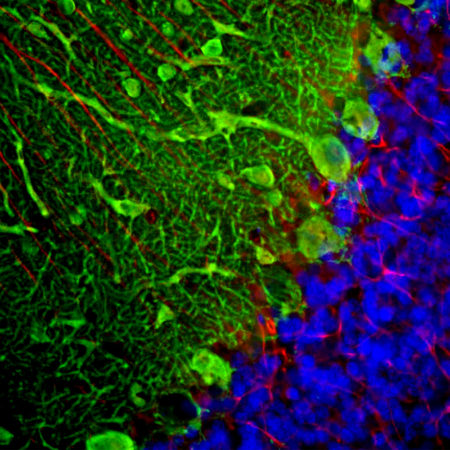
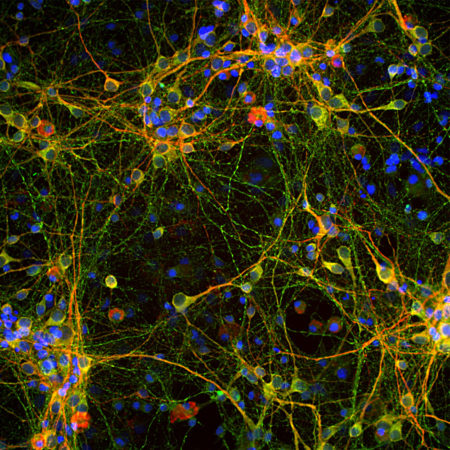
Confocal immunofluorescent analysis of HeLa cells stained with chicken pAb to fibrillarin, CPCA-fib, dilution 1:10,000 in green and costained with mouse mAb to vimentin, MCA-2D1, 1:1,000, in red. The blue signal is DAPI staining of nuclear DNA. The fibrillarin antibody stains nucleoli while the vimentin antibody binds to cytoplasmic intermediate filaments.
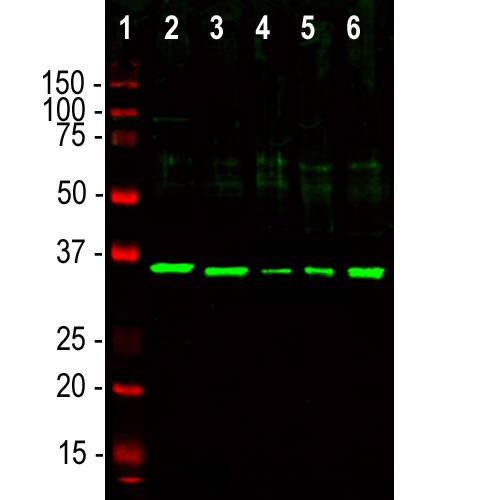
Western blot analysis of different cell lysates using chicken pAb to fibrillarin CPCA-Fib, dilution 1:5,000, in green: [1] protein standard (red), [2] NIH-3T3, [3] HEK293, [4] HeLa, [5] SH-SY5Y, and [6] C6 cells. The single strong band at ~35kDa correspond to the fibrillarin protein.
Chicken Polyclonal Antibody to Fibrillarin
Cat# CPCA-Fib
$120.00 – $800.00
Fibrillarin is a highly conserved component of a nucleolar small ribonucleoprotein complex in mammals, involved in the processing of ribosomal RNA during ribosomal biogenesis. The protein runs at ~35kDa on SDS-PAGE and is very rich in basic amino acids having a PI of 9.8. Fibrillarin was originally identified in humans since autoantibodies staining nucleoli were seen in some patients with the autoimmune disease scleroderma (1). Subsequently the protein fibrillarin was found to be the human homologue of Nop1p, a Saccharomyces cerevisiae nucleolar protein, the two proteins being 67% identical (2,3). We have generated an alignment of the sequences of fibrillarin and homologues downloadable from here. The fibrillarin molecule consists of an N-terminal glycine and arginine rich region followed by a highly conserved globular domain. Embryonic knockout of the fibrillarin gene in mice is lethal, suggesting fundamental importance of this protein (4). Autoantibodies to fibrillarin are also seen in patients with the autoimmune disease systemic sclerocis (5).
The CPCA-Fib antibody was made against recombinant human fibrillarin expressed in and purified from E. coli and is superior on western blots of mammalian samples to the widely used MCA-38F3 antibody, which was originally raised against yeast Nop1p and later found to recognize fibrillarin, the mammalian homologue of the yeast protein. However MCA-38F3 has been documented to be usable as a marker of nucleoli in a wide variety of species which has not so far been documented with this reagent. The chicken antibody works well for IF and ICC but is not recommended for IHC. We have also produced a rabbit polyclonal antibody to fibrillarin RPCA-Fib and an alternate mouse monoclonal MCA-4A4, both made against recombinant human fibrillarin. Mouse select image above left for larger view.
Nop1p was originally identified as a nucleolar protein of bakers yeast, Saccharomyces cerevisiae (accession P15646). The Nop1p protein is 327 amino acids in size (34.5kDa), is essential for yeast viability, and is localized in the nucleoli (1). The systematic name for S. cerevisiae Nop1 is YDL014W, and it is now known to be part of the small subunit processome complex, involved in the processing of pre-18S ribosomal RNA. Nop1p is the yeast homologue of a protein apparently found in all eukaryotes and archea generally called fibrillarin. Fibrillarin/Nop1p is extraordinarily conserved, so that the yeast and human proteins are 67% identical, and the human protein can functionally replace the yeast protein. An alignment of fibrillarin and homologues from a variety of species showing the high cross species sequence conservation: download from here. This means that suitably cross-reactive antibodies to Nop1p/fibrillarin, like MCA-4A4, can be used to reveal nucleoli and study fibrillarin/Nop1p in all eukaryotes and archea tested to date.
Human fibrillarin has been characterized (accession P22087) and the human fibrillarin gene is located on chromosome 19 (19q13.1). Fibrillarin/Nop1p proteins have been cloned and sequenced from several other species (e.g. Mouse, accession P35550, Xenopus accession P22232, C. elegans accession Q22053, and S. pombe accession P35551. The N terminal ~80 amino acids contain multiple copies based on the peptide RGG, or arginine-glycine-glycine, sometimes referred to as GAR repeats, characteristic of the GAR family of molecules. The remaining ~240 amino acids consist of the so called fibrillarin domain. A fibrillarin homologue has also been identified in the genome of the archean Methanococcus (accession NC_000909). This protein lacks the RGG rich N-terminal extension but is clearly homologous to the other sequences throughout all of the fibrillarin domain. The 3D structure of this molecule has been determined and shown to consist of 2 extended β-sheets flanked by α-helixes (Medline link). Patients with the autoimmune disease scleroderma often have strong circulating autoantibodies to a ~34kDa protein which was subsequently found to be fibrillarin. Recent studies show that knock out of the fibrillarin gene in mice results in embryonic lethality, although mice with only one functional fibrillarin/Nop1p gene were viable (3).
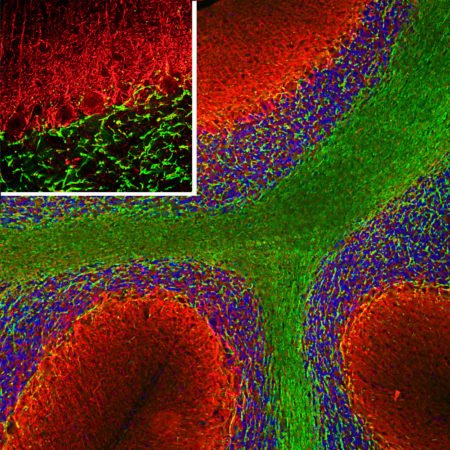
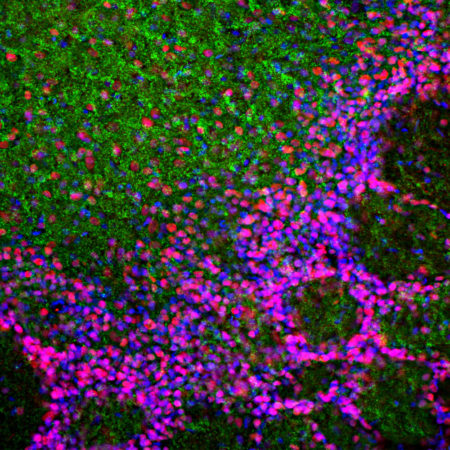
Fibrillarin/Nop1p antibody has become widely used as a convenient marker for nucleoli in a wide variety of species (e.g. 4-6). The HGNC name for this protein is FBL. To raise the CPCA-Fib antibody, mice were injected with full length recombinant human fibrillarin.
An alignment of fibrillarin and homologues from a variety of species showing the high cross species sequence conservation: download from here.
1. Aris JP and Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell Biol. 107:17-31 (1988).
2. Aris JP and Blobel G. cDNA cloning and sequencing of human fibrillarin, a conserved nucleolar protein recognized by autoimmune antisera. Proc. Natl. Acad. Sci. 88:931-5 (1991).
3. Ochs RL, Lischwe MA, Spohn WH, Busch H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol. Cell. 54:123-33 (1985).
4. Newton K, Petfalski E, Tollervey D, Caceres JF. Fibrillarin is essential for early development and required for accumulation of an intron-encoded small nucleolar RNA in the mouse. Mol. Cell Biol. 23:8519-27 (2003).
5. Okano Y, Steen VD, Medsger TA. Autoantibody to U3 nucleolar ribonucleoprotein (fibrillarin) in patients with systemic sclerosis. Arth. Rheum. 35:95-100 (1992).
Related products
Contact info
EnCor Biotechnology Inc.
4949 SW 41st Boulevard, Ste 40
Gainesville
Florida 32608 USA
Phone: (352) 372 7022
Fax: (352) 372 7066
E-mail: admin@encorbio.com