| Name: | Mouse Monoclonal to Muscleblind like protein 1. |
| Immunogen: | Full-length recombinant human muscleblind-like 1 protein expressed in and purified from E. coli |
| HGNC Name: | MBNL1 |
| UniProt: | Q9NR56 |
| Molecular Weight: | 39kDa |
| Host: | Mouse |
| Isotype: | IgG1 |
| Species Cross-Reactivity: | Human, no reactivity with rat or mouse |
| RRID: | AB_2572351 |
| Format: | Purified antibody at 1mg/mL in 50% PBS, 50% glycerol plus 5mM NaN3 |
| Applications: | WB, IF/ICC |
| Recommended Dilutions: | WB: 1:1,000-1:2,000. ICC/IF: 1:1,000. IHC: not recommended |
| Storage: | Store at 4°C for short term, for longer term at -20°C. |
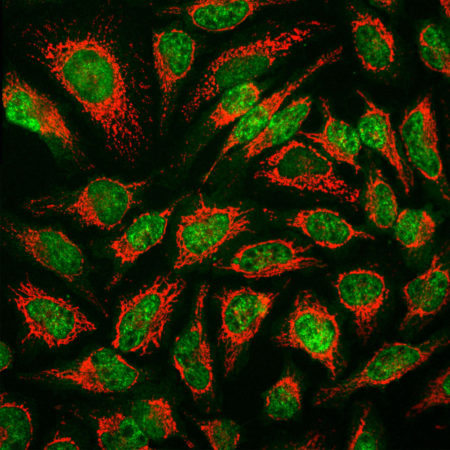
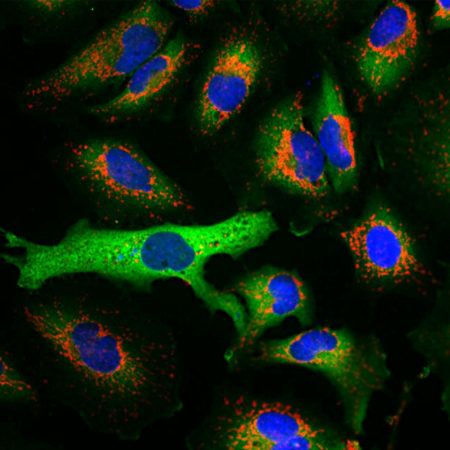
Immunofluorescent analysis of HeLa cells stained with mouse mAb to MBNL1, MCA-1H1, dilution 1:1,000 in green and chicken pAb to HSP60, CPCA-HSP60, dilution 1:5,000, in red. The MCA-1H1 antibody specifically labels nuclei, while the CPCA-HSP60 antibody reveals protein expressed in mitochondria.
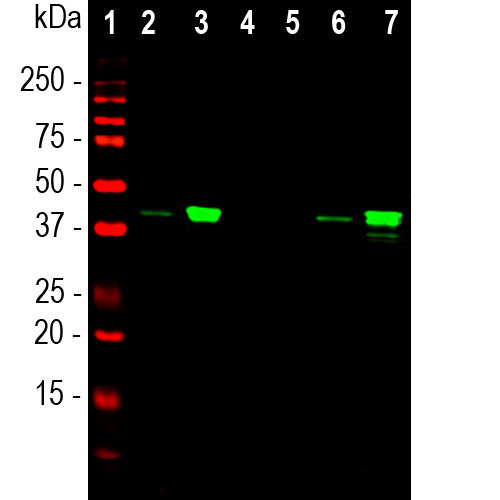
Western blot analysis of whole cell lysates, cytosol or nuclear enriched fractions, using mouse mAb to MBNL1, MCA-1H1, dilution 1:1,000 in green: [1] protein standard (red), [2] HEK293 cytosol, [3] HEK293 nuclear fraction, [4] NIH-3T3 cytosol, [5] NIH-3T3 nuclear fraction, [6] HeLa cytosol, and [7] HeLa nuclear fraction. The strong band at the 40kDa corresponds to the MBNL1 protein that is predominantly detected in the nuclear enriched fractions. This antibody binds exclusively to the human protein and does not bind the rodent homologue.
Mouse Monoclonal Antibody to MBNL1
Cat# MCA-1H1
$120.00 – $800.00
A protein named “muscleblind” was originally isolated following studies of Drosophila, since inactivation of the muscleblind gene in this species resulted in defects the development of both muscles and the visual system (1). Subsequently gene cloning identified three mammalian muscleblind like protein homologues were discovered, MBNL1 (a.k.a. MBNL), MBNL2 (a.k.a. MBLL) and MBNL3 (a.k.a. MBXL/MBLX/CHCR). The MBNL1 protein was also discovered since it binds specifically to the aberrant polynucleotide repeats derived from the myotonin gene which are causative of myotonic dystrophy type 1 (DM1). This is one of the diseases in which the copy number of 3-6 base DNA repeats increases beyond the normal level and cause clinical problems (2-4). In the case of DM1 the 3′ UTR of the myotonin gene normally has 5-37 copies of a CTG trinucleotide repeat which in the disease state is increased to 50-5000 repeats. These repeated sequences in the mRNA bind to and sequester the MBNL1 protein. The myotonin gene codes for a serine/threonine protein kinase predominantly expressed in muscle, and which is related in sequence to other protein kinases regulated by small G proteins. The three muscle blind-like proteins have differing expression patterns in mice, with MBNL1 being expressed widely, with particularly high levels in heart, muscle and liver (5). The MBNL1 protein localizes on nuclear foci in cells derived from DM1 patients along with the repeat containing mRNA (6,7). Knockout of MBNL1 in mice leads to muscle and eye deficits similar to those seen in Drosophila (8). The mice also showed RNA splicing abnormalities that are characteristic of the DM1 disease process, and it is thought that MBNL1 therefore has a role in the regulation of alternate mRNA splicing.
The MCA-1H1 antibody was made against full length recombinant human MBNL1 expressed in and purified from E. coli. It binds human MBNL1 on western blots and for IF but does not recognize the rat or mouse MBNL1 homologue, so is not recommended for studies of rodents. It is also not recommended for IHC. The antibody is also a useful marker of the nuclear fraction in biochemical fractionation experiments. Mouse select image at left for larger view.
1. Begemann G, et al. Muscleblind, a gene required for photoreceptor differentiation in Drosophila, encodes novel nuclear Cys3His-type Zinc-finger-containing proteins. Development 124:4321-31 (1997).
2. Jansen G et al. Abnormal myotonic dystrophy protein kinase levels produce only mild myopathy in mice. Nat. Genet. 13:316-24 (1996).
3. Reddy S et al. Mice lacking the myotonic dystrophy protein kinase develop a late onset progressive myopathy. Nat. Genet. 13:325-35 (1996).
4. Miller JW, et al. Recruitment of human muscleblind proteins to (CUG)n expansions associated with myotonic dystrophy. EMBO J. 19:4439-48 (2000).
5. Kanadia RN, et al. Developmental expression of mouse muscleblind genes Mbnl1, Mbnl2 and Mbnl3. Gene Expr. Patterns 3:459-62 (2003).
6. Fardaei M, et al. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet. 11:805-14 (2002).
7. Ho TH, et al. Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J. Cell Sci. 118:2923-33 (2005).
8. Kanadia RN, et al. A muscleblind knockout model for myotonic dystrophy. Science 302:1978-80 (2003).
Related products
Contact info
EnCor Biotechnology Inc.
4949 SW 41st Boulevard, Ste 40
Gainesville
Florida 32608 USA
Phone: (352) 372 7022
Fax: (352) 372 7066
E-mail: admin@encorbio.com