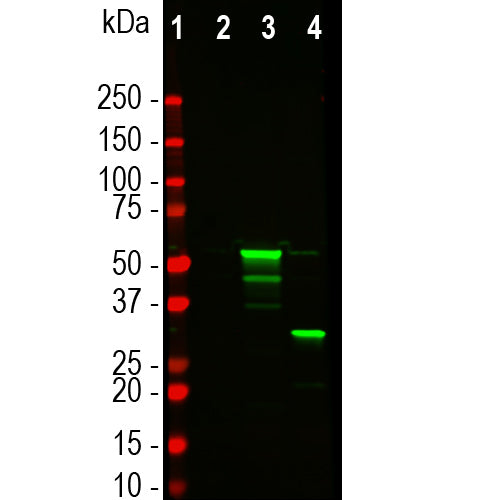
![Western blot analysis of HEK293 cell lysates, using mouse mAb to tdTomato, MCA-6F12, dilution 1:2,000, in green [1] protein standard, [2] untransfected HEK293 lysate, [3] HEK293 cells transfected with pCI-Neo-mod vector expressing tdTomato protein, and [4] HEK293 transfected with mCherry-HA construct. Major band at about 60kDa corresponds to the tdTomato protein which contains two fluorescent domains.](http://encorbio.com/cdn/shop/files/MCA-6F12_Tomato_WBTF_HEK_ct_tom_cher_20x_crop_center.jpg?v=1759251164)
EnCor Biotechnology
Mouse Monoclonal Antibody to tdTomato (Tomato), Cat# MCA-6F12
Description
The MCA-6F12 antibody was made against full length recombinant tdTomato expressed in and purified from E. coli. The tdTomato protein contains two fluorescent Tomato units connected by a Ser and Gly rich linker sequence. This has superior spectral properties to a monomeric form and is included in many widely used expression vectors, such as WOZ44746.1. The antibody recognizes tdTomato strongly on western blots, in appropriate transfected and transgenic cells and tissues and does not react with GFP. The tdTomato protein was derived from DSRed, a red fluorescent protein originally discovered in the coral Discosoma. The MCA-6F12 antibody also reacts with the closely related protein mCherry, also derived from DSRed. The epitope has been localized to amino acids 151-170 of mCherry, corresponding to 147-166 of tdTomato. All these proteins are similar in size and general structure to GFP, though distinct in primary sequence and spectral properties (5,6).
- Epitope Mapped Antibodies
- Fluorescent Protein Antibodies
- Immunohistochemistry Verified
- Mouse Monoclonal Antibodies
- New Antibodies
Add a short description for this tabbed section
| Immunogen: | Full length two domain recombinant tdTomato protein |
| UniProt: | D1MPT3 |
| Molecular Weight: | ~56kDa |
| Host: | Mouse |
| Isotype: | IgG1 |
| Species Cross-Reactivity: | N.A. |
| Format: | Protein G affinity purified antibody at 1mg/mL in 50% PBS, 50% glycerol plus 5mM NaN3 |
| Applications: | WB, IF/ICC, IHC |
| Recommended Dilutions: | WB: 1:2,000. IF/IHC: 1:1000-1:2,000. |
| Storage: | Store at 4°C for short term, for longer term store at -20°C. Stable for 12 months from date of receipt. |
The Tomato fluorescent protein is derived from a natural product, DsRed, originally isolated as a red fluorescent protein from the coral of the genus Discosoma (1). Most vectors incorporating Tomato protein express two back to back copies, and this form is therefore referred to as "tandem dimer Tomato" or tdTomato. As with other natural fluorescent proteins of Cnidarians (jelly fish, sea anemones and corals), the natural form of the protein forms stable tetramers in vivo. DsRed, the original form, was engineered to improve its spectral properties and also prevent multimerization in the lab of Roger Tsien, where much work on fluorescent proteins was performed (2). Several further cycles of mutation, directed modification and evolutionary selection produced dTomato, which has an excitation maximum at 554nm and and emission maximum at 581nm (3). The protein is widely used as a fluorescent tracer in transfection, transgenic, photobleaching and FRET type experiments. The prototype for these fluorescent proteins is Green Fluorescent Protein (GFP), which is a ~27kDa protein isolated originally from the jellyfish Aequoria victoria (4). The Tomato protein is similar in size and general structural properties to GFP (5,6), but, obviously, produces a red rather than a green fluorochrome. As with GFP, Tomato becomes fluorescent due to intrinsic properties requiring only molecular oxygen and so can be readily expressed in a variety of systems.
This antibody cross reacts with the related mCherry protein, unsurprising since both proteins were derived from DSRed. We tested binding of this antibody to recombinant tdTomato in the presence of a set of nested 20 amino acid peptides staggered by 5 amino acids. One mCherry peptide, 151-170, SSERMYPEDGALKGEIKQRL showed very strong inhibition. The corresponding peptide in tdTomato is the related STERLYPRDGVLKGEIHQAL, amino acids 147-166.
This antibody was made against a recombinant tdTomato construct expressed in and purified from E. coli. The sequence is identical to that found in a series of widely used expression vectors, including two Tomato fluorescent units linked by a glycine and serine rich linker. The linker in the sequence below is underlined. The protein was expressed in the eukaryotic expression vector pET30a(+) which adds an N terminal His-tag and some other sequence, underlined below. This sequence includes a thrombin cleavage site (blue), an S-tag affinity peptide (red) and an enterokinase cleavage site (green). The sequence is identical to that in the WOZ44746.1 expression vector.
MHHHHHHSSG LVPRGSGMKE TAAAKFERQH MDSPDLGTDD DDKAMADIGS EFMVSKGEEV
IKEFMRFKVR MEGSMNGHEF EIEGEGEGRP YEGTQTAKLK VTKGGPLPFA WDILSPQFMY
GSKAYVKHPA DIPDYKKLSF PEGFKWERVM NFEDGGLVTV TQDSSLQDGT LIYKVKMRGT
NFPPDGPVMQ KKTMGWEAST ERLYPRDGVL KGEIHQALKL KDGGHYLVEF KTIYMAKKPV
QLPGYYYVDT KLDITSHNED YTIVEQYERS EGRHHLFLGH GTGSTGSGSS GTASSEDNNM
AVIKEFMRFK VRMEGSMNGH EFEIEGEGEG RPYEGTQTAK LKVTKGGPLP FAWDILSPQF
MYGSKAYVKH PADIPDYKKL SFPEGFKWER VMNFEDGGLV TVTQDSSLQD GTLIYKVKMR
GTNFPPDGPV MQKKTMGWEA STERLYPRDG VLKGEIHQAL KLKDGGHYLV EFKTIYMAKK
PVQLPGYYYV DTKLDITSHN EDYTIVEQYE RSEGRHHLFL YGMDELYK
Number of amino acids: 528
Molecular weight: 59903.76
Theoretical pI: 6.08
Amino acid composition:
Ala (A) 21 4.0%
Arg (R) 20 3.8%
Asn (N) 10 1.9%
Asp (D) 33 6.2%
Cys (C) 0 0.0%
Gln (Q) 17 3.2%
Glu (E) 45 8.5%
Gly (G) 57 10.8%
His (H) 22 4.2%
Ile (I) 19 3.6%
Leu (L) 35 6.6%
Lys (K) 47 8.9%
Met (M) 25 4.7%
Phe (F) 24 4.5%
Pro (P) 28 5.3%
Ser (S) 30 5.7%
Thr (T) 31 5.9%
Trp (W) 6 1.1%
Tyr (Y) 28 5.3%
Val (V) 30 5.7%
Total number of negatively charged residues (Asp + Glu): 78
Total number of positively charged residues (Arg + Lys): 67
Extinction coefficients:
Extinction coefficients are in units of M-1 cm-1, at 280 nm measured in water.
Ext. coefficient 74720
Abs 0.1% (=1 g/l) 1.247
1. Matz MV, et al. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 17:969-73 (1999).
2. Baird GS, Zacharias DA, Tsien RY. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. PNAS 97:11984-9 (2000).
3. Chalfie M, et al. Green fluorescent protein as a marker for gene expression. Science 263:802-5 (1994).
4. Gross LA. et al. The structure of the chromophore within DsRed, a red fluorescent protein from coral. PNAS 97:11990-5 (2000).
5. Heikal AA. et al. Molecular spectroscopy and dynamics of intrinsically fluorescent proteins: coral red (dsRed) and yellow (Citrine). PNAS 97:11996-2001 (2000).
6. Shaner NC. et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotech. 22:1567-72 (2004).
Add a short description for this tabbed section