EnCor Biotechnology
Mouse Monoclonal Antibody to SARS-CoV2 S-Protein ACE2 Binding Domain, Cat# MCA-2G1
Description
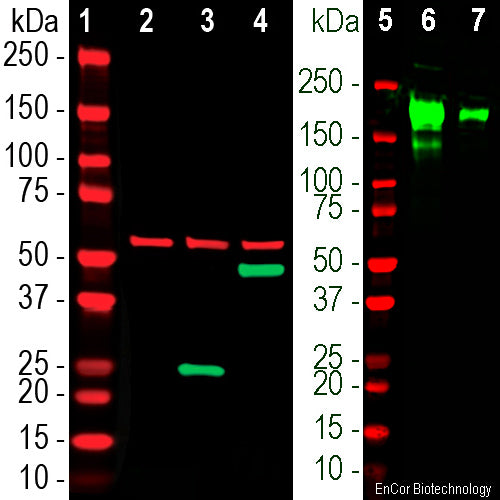
The MCA-2G1 antibody was made against our recombinant construct comprising amino acids 308-541 in the S-protein sequence in SARS-CoV2 Wuhan-Hu-1, complete genome. The antibody works well on western blots of crude homogenates of HEK293 cells transfected with the SARS-CoV2 binding domain, cleanly producing the appropriate sized band and as expected also binds the full length S-protein. In addition S-protein transfected cells show clean and strong immunofluorescence staining of the expressed protein with this antibody. We are currently determining the exact peptide epitope of this and our other SARS-CoV2 S-protein antibodies and also measuring their kinetic properties. EnCor supplies another mouse monoclonal antibody to the SARS-CoV2 S-protein ACE2 binding domain MCA-5G8 and also a rabbit polyclonal RPCA-SARS-CoV2-bd.
Add a short description for this tabbed section
| Immunogen: | Recombinant SARS-CoV2 S-Protein ACE2 binding domain expressed in and purified from E. coli, EnCor product PROT-r-SARS-CoV2-bd |
| HGNC Name: | N.A. |
| UniProt: | P0DTC2 |
| Molecular Weight: | S-Protein 142kDa |
| Host: | Mouse |
| Isotype: | IgG1 |
| Species Cross-Reactivity: | N.A. |
| RRID: | AB_2861173 |
| Format: | Protein G affinity purified antibody at 1mg/mL in 50% PBS, 50% glycerol plus 5mM NaN3 |
| Applications: | WB, ICC/IF |
| Recommended Dilutions: | WB: 1:1,000-1:3,000. ICC/IF: 1:1,000. IHC: Not Tested. |
| Storage: | Store at 4°C for short term, for longer term store at -20°C. Stable for 12 months from date of receipt. |
In late 2019 a novel infectious disease was discovered in Wuhan, China which was quickly recognized to be caused by a previously unknown RNA coronavirus. The virus was very rapidly isolated, the full RNA sequence determined and put on-line on the 10th of January 2020. The sequence revealed that the virus was most closely related to certain bat coronaviruses and the severe acute respiratory syndrome (SARS) coronavirus. Immediately biotechnology companies and research institutes used the RNA sequence information to generate vaccine candidates. The SARS virus was known to enter and infect human cells by means of the so-called spike or S-protein which binds to the extracellular domain of the angiotensin converting enzyme 2 (ACE2) protein, which is then internalized bringing the virus into the cell. Cryoelectron microscopy and binding studies quickly determined that the S-protein of SARS-CoV2 is structurally similar to to that of the SARS virus and also binds to the ACE2 receptor, albeit with higher affinity than the S-protein of SARS. This focuses attention on the ACE2 binding site on the SARS-CoV2 S-protein and for the complementary region on ACE2 which binds the SARS-CoV2 S-protein. We therefore expressed both these regions in E. coli, our products PROT-R-SARS-CoV2-bd and PROT-R-ACE2-bd and raised antibodies to them.

Vero cells are a cell line derived derived from monkey kidney and express an ACE2 receptor similar to that found in humans, and are therefore susceptible to infection by patient derived SARS-CoV2 virus. The cells were grown in culture, fixed, and stained with the DNA dye DAPI, revealing cell nuclei (control, top left), The remaining three panels show staining of similar cells following infection with SARS-CoV2 virus. Cells were then reacted with all three EnCor antibodies to the cell binding domain of the SARS-CoV2 spike protein, namely MCA-2G1, MCA-5G8 and RPCA-SARS-CoV2-bd as indicated. Antibody binding was revealed with appropriate ALEXA green (top right, bottom left) or red (bottom right) secondary antibodies. The virus accumulates in the cytoplasm of infected cells. The cells were grown and infected under appropriate safety and containment in the University of Florida Department of Pathology. Mouse select image above for larger view.

Immunofluorescent analysis of transfected HEK293 cells with the SARS-CoV2-bd-construct under high magnification, stained with mouse mAb to SARS-CoV2-bd, MCA-2G1, dilution 1:1,000, in green. Cells were costained with rabbit pAb to HSP60, RPCA-HSP60, dilution 1:2,000, in red. The blue is Hoechst staining of nuclear DNA. MCA-2G1 antibody reveals overexpression of SARS-CoV2-bd protein presumanbly in lysosomes only in transfected cells, while the HSP60 antibody labels mitochondria in all cells. Mouse select image above left for larger view.
This antibody was raised against a recombinant construct of the SARS-CoV-2 spike or S-protein which includes the entire region which interacts with ACE2. The specific binding to ACE2 is essential for viral internalization and infection. We designed this construct based on amino acids 308-541 in the S-protein sequence in Isolate Wuhan-Hu-1, complete genome. This is a defined globular domain recently shown to include all of the amino acids necessary for ACE2 binding. The construct was expressed in and purified from E. coli and includes an N-terminal His-tag and other vector derived sequence shown underlined below. Amino acids which interact directly with the ACE2 protein are printed in bold, data based on the cryoEM study of Walls et al. (3).
MHHHHHHSSG LVPRGSGMKE TAAAKFERQH MDSPDLGTDD DDKAMADIGS EFVEKGIYQT 60
SNFRVQPTES IVRFPNITNL CPFGEVFNAT RFASVYAWNR KRISNCVADY SVLYNSASFS 120
TFKCYGVSPT KLNDLCFTNV YADSFVIRGD EVRQIAPGQT GKIADYNYKL PDDFTGCVIA 180
WNSNNLDSKV GGNYNYLYRL FRKSNLKPFE RDISTEIYQA GSTPCNGVEG FNCYFPLQSY 240
GFQPTNGVGY QPYRVVVLSF ELLHAPATVC GPKKSTNLVK NKCVNF 286
Number of amino acids: 286
Molecular weight: 32074.01
Theoretical pI: 8.01
Amino acid composition:
Ala (A) 17 5.9%
Arg (R) 13 4.5%
Asn (N) 22 7.7%
Asp (D) 16 5.6%
Cys (C) 9 3.1%
Gln (Q) 9 3.1%
Glu (E) 11 3.8%
Gly (G) 21 7.3%
His (H) 8 2.8%
Ile (I) 11 3.8%
Leu (L) 16 5.6%
Lys (K) 16 5.6%
Met (M) 4 1.4%
Phe (F) 19 6.6%
Pro (P) 15 5.2%
Ser (S) 23 8.0%
Thr (T) 16 5.6%
Trp (W) 2 0.7%
Tyr (Y) 16 5.6%
Val (V) 22 7.7%
Total number of negatively charged residues (Asp + Glu): 27
Total number of positively charged residues (Arg + Lys): 29
Extinction coefficient 35340
Abs 0.1% (=1 g/l) 1.102, assuming all pairs of Cys residues form cystines
Extinction coefficient 34840
Abs 0.1% (=1 g/l) 1.086, assuming all Cys residues are reduced
This antibody has not been tested on formalin fixed and paraffin embedded samples for IHC, and iso we cannot recommended it for this purpose.
1. Wu, F et al. A new coronavirus associated with human respiratory disease in China. Nature doi:10.1038/s41586-020-2008-3.2020 579:265-269 (2020).
2. Ren, L-L et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J (Engl) doi:10.1097/CM9.0000000000000722 133:1015-24 (2020).
3. Walls, A C et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell doi: 10.1016/j.cell.2020.02.058 180:1-12 (2020)
4. Yan, R et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science doi:10.1126/science.abb2762 367:1444–8 (2020).
5. Wang, D-S et al. The pleckstrin homology domain of human β-I σ-II spectrin is targeted to the plasma membrane in vivo. Biochem. Biophys. Res. Comm. 225:420-6 (1996).
Add a short description for this tabbed section