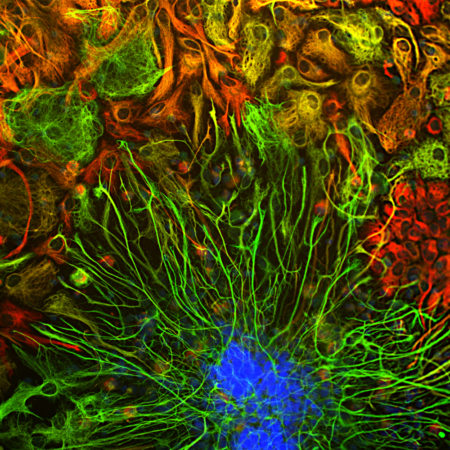
Fluorescent proteins have become widely used in a variety of experimental paradigms since the characterization of the first one, Green Fluorescent Protein (GFP) about 20 years ago. Some of these fluorescent proteins have the useful property of changing their emission spectrum from green to red following irradiation with blue or UV light, which allows simple but powerful pulse chase type experiments to be performed using more or standard fluorescence microscopes. We have expressed in E. coli an purified a protein originally isolated from Lobophyllia hemprichii, a type of stony coral. The protein was described in Wiedenannn et al. (2004) and named EosFP, after Eos, the goddess of dawn in Greek mythology. Upon appropriate irradiation the protein changes emission from 516 nm to 581 nm, which happens to fit very conveniently to typical green (~498 nm) and red (~594 nm) filters on fluorescence microscopes. The emission shift is due to an irreversible covalent modification in the fluorochrome and is dependent on the His residue in the His-Tyr-Gly sequence that produces the fluorescence. The original coral protein was mutagenized to prevent tetramerization and dimerization, a requirement if the protein is to be used for fusion protein generation useful for FRET and similar techniques. A recent advance was the development of CaMPARI, an acronym for “Calcium-Modulated Photoactivatable Ratiometric Integrator” (2). This construct contains the GCaMP Calcium indicator fused to two EosFP domains, and will only transit from green to red when there is a coincidence between Calcium level elevation and the appropriate wavelength of light. This allows functional mapping of cellular activation in real time in appropriate transgenic animals (2).
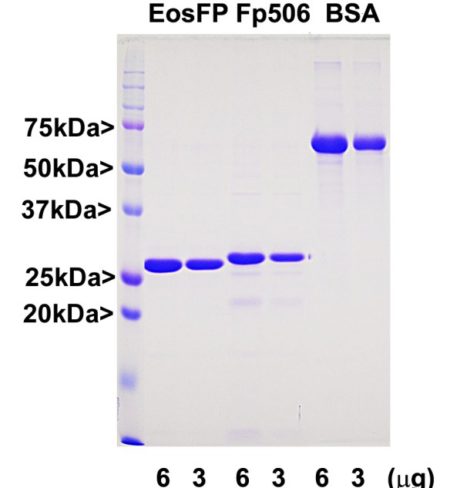
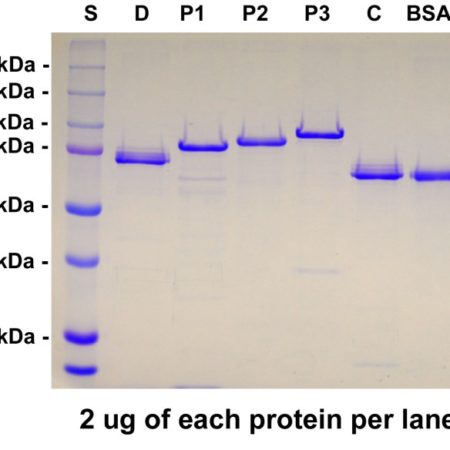
Our EosFP construct is identical to that in GenPet database entry AAV54099, as described in reference 1. The vector adds a C-terminal His-tag which was use to purify the protein and this, along with some other vector derived sequence, adds about 5 kDa to the molecule.
Fluorescent proteins have become widely used in a variety of experimental paradigms since the characterization of the first one, Green Fluorescent Protein (GFP) about 20 years ago. Some of these fluorescent proteins have the useful property of changing their emission spectrum from green to red following irradiation with blue or UV light, which allows simple but powerful pulse chase type experiments to be performed using more or standard fluorescence microscopes. We have expressed in E. coli an purified a protein originally isolated from Lobophyllia hemprichii, a type of stony coral. The protein was described in Wiedenannn et al. (2004) and named EosFP, after Eos, the goddess of dawn in Greek mythology. Upon appropriate irradiation the protein changes emission from 516 nm to 581 nm, which happens to fit very conveniently to typical green (~498 nm) and red (~594 nm) filters on fluorescence microscopes. The emission shift is due to an irreversible covalent modification in the fluorochrome and is dependent on the His residue in the His-Tyr-Gly sequence that produces the fluorescence. The original coral protein was mutagenized to prevent tetramerization and dimerization, a requirement if the protein is to be used for fusion protein generation useful for FRET and similar techniques. A recent advance was the development of CaMPARI, an acronym for “Calcium-Modulated Photoactivatable Ratiometric Integrator” (2). This construct contains the GCaMP Calcium indicator fused to two EosFP domains, and will only transit from green to red when there is a coincidence between Calcium level elevation and the appropriate wavelength of light. This allows functional mapping of cellular activation in real time in appropriate transgenic animals (2).
Our EosFP construct is identical to that in GenPet database entry AAV54099, as described in reference 1. The vector adds a C-terminal His-tag which was use to purify the protein and this, along with some other vector derived sequence, adds about 5 kDa to the molecule.