EnCor Biotechnology
Goat Polyclonal Antibody to mCherry, Cat# GPCA-mCherry
Description
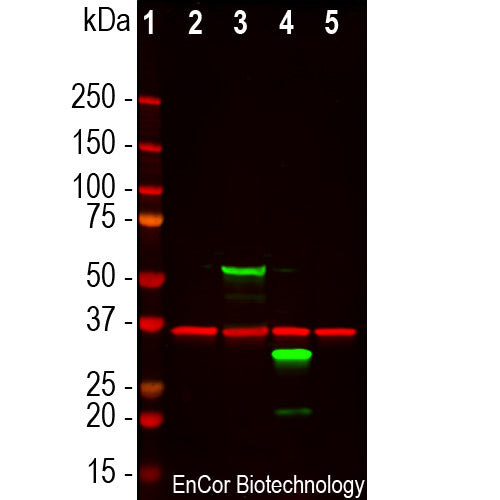
GPCA-mCherry antibody was made against full length recombinant mCherry protein expressed in and purified from E. coli, Prot-r-mCherry. The antibody recognizes mCherry and tdTomato on western blots and in appropriate cells and sections. It does not react with GFP. GPCA-mCherry antibody can be used to verify the size of fusion constructs by western blotting, and to amplify the endogenous fluorescence of mCherry in transfected cells. We also supply a mouse monoclonal antibody to mCherry, MCA-1C51, and MCA-5A6, as well as goat and chicken polyclonal antibodies to this protein, GPCA-mCherry and CPCA-mCherry respectively.
- Fluorescent Protein Antibodies
- Goat Polyclonal Antibodies
- Immunohistochemistry Verified
- Our Most Widely Used Reagents
Add a short description for this tabbed section
| Immunogen: | Full length recombinant mCherry protein |
| HGNC Name: | N.A. |
| UniProt: | D1MPT3 |
| Molecular Weight: | ~28kDa |
| Host: | Goat |
| Species Cross-Reactivity: | N.A. |
| RRID: | AB_2858265 |
| Format: | Immunogen affinity purified antibody at 1mg/mL in 50% PBS, 50% glycerol plus 5mM NaN3 |
| Applications: | WB, IF/ICC, IHC |
| Recommended Dilutions: | WB: 1:1,000. ICC 1:2,000. IF/IHC: 1:5,000-1:10,000. |
| Storage: | Store at 4°C for short term, for longer term at -20°C. Stable for 12 months from date of receipt. |
The mCherry protein is engineered from a fluorescent protein originally isolated from a coral and is widely used as a tracer in transfection and transgenic experiments. The prototype for these fluorescent proteins is Green Fluorescent Protein (GFP), which is a ~27kDa protein isolated originally from the jellyfish Aequoria victoria. GFP was the basis of the 2008 Nobel Prize in Chemistry, awarded to Osamu Shimomura, Martin Chalfie and Roger Tsien, specifically “for the discovery and development of the green fluorescent protein, GFP”. The mCherry protein is derived from DsRed, a red fluorescent protein related to GFP isolated from disc corals of the genus Discosoma. DsRed is similar in size and properties to GFP, but, obviously, produces a red rather than a green fluorochrome. The original DsRed was engineered extensively in the Tsien lab to prevent it from forming tetramers and dimers and to modify and improve the spectral properties (1-3). Several further cycles of mutation, directed modification and evolutionary selection produced mCherry, which has an excitation maximum at 587nm and and emission maximum at 610nm (4).Several further cycles of mutation, directed modification and evolutionary selection produced mCherry, which has an excitation maximum at 587nm and and emission maximum at 610nm (4). The same lab engineered other fluorescent DsRed derivatives such as tdTomato, mOrange, mStrawberry and others. This antibody likely binds all these variants and is known to bind tdTomato.

Chromogenic immunostaining of a 4% PFA fixed paraffin embedded section of mouse brain transduced with a TdTomato construct under an oxytocin promoter. Staining with goat pAb to mCherry, GPCA-mCherry, dilution 1:10,000 detected with DAB (brown) using the ABC method with citrate buffer retrieval at pH=6.0. Hematoxylin (blue) was used as the counterstain. GPCA-mCherry specifically detected the soma and axons of oxytocin positive neurons in the hypothalamus. The TdTomato protein is very similar in sequence to mCherry so that our antibody strongly recognizes both proteins. Mouse select image for larger view.
This antibody was made against a recombinant construct expressed in and purified from E. coli. The sequence is identical to that found in a series of widely used expression vectors and is identical to Uniprot entry D1MPT3.
1 MVSKGEEDNM AIIKEFMRFK VHMEGSVNGH EFEIEGEGEG RPYEGTQTAK
51 LKVTKGGPLP FAWDILSPQF MYGSKAYVKH PADIPDYLKL SFPEGFKWER
101 VMNFEDGGVV TVTQDSSLQD GEFIYKVKLR GTNFPSDGPV MQKKTMGWEA
151 SSERMYPEDG ALKGEIKQRL KLKDGGHYDA EVKTTYKAKK PVQLPGAYNV
201 NIKLDITSHN EDYTIVEQYE RAEGRHSTGG MDELYK
1. Baird GS, Zacharias DA, Tsien RY. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. PNAS 97:11984-9 (2000).
2. Gross LA, et al. The structure of the chromophore within DsRed, a red fluorescent protein from coral. PNAS 97:11990-5 (2000).
3. Heikal AA, et al. Molecular spectroscopy and dynamics of intrinsically fluorescent proteins: coral red (dsRed) and yellow (Citrine). PNAS 97:11996-2001 (2000).
4. Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotech. 22:1567-72 (2004).
Add a short description for this tabbed section