EnCor Biotechnology
Rabbit Polyclonal Antibody to Cas-φ/Cas12j, Cat# RPCA-Cas12j
Description
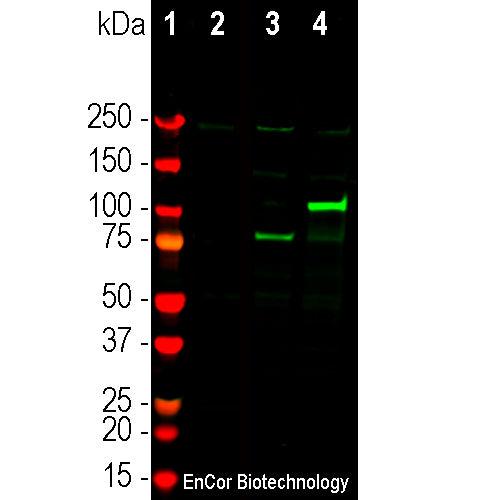
The RPCA-Cas12j antibody was made against recombinant construct of the full sequence of Casφ-2 from Pausch et al. 2020, derived from the amusingly named Biggievirus Mos11. The antibody works well on western blots of cell lysates of HEK293 cells transfected with the Casφ-2 DNA, cleanly producing the expected sized band. In addition such transfected cells show clean and strong cytoplasmic staining by immunofluorescence methods with this antibody. We also supply a mouse monoclonal antibody to this protein, MCA-5F95.
Add a short description for this tabbed section
| Immunogen: | Full length CasΦ-2 expressed in and purified from E. coli. |
| HGNC Name: | N.A. |
| UniProt: | N.A. |
| Molecular Weight: | -90kDa |
| Host: | Rabbit |
| Species Cross-Reactivity: | N.A. |
| RRID: | 2889160 |
| Format: | Purified antibody at 1mg/mL in 50% PBS, 50% glycerol plus 5mM NaN3 |
| Applications: | WB, IF/ICC |
| Recommended Dilutions: | WB: 1:1,000-2,000. IF/ICC: 1:2,000-5,000. |
| Storage: | Store at 4°C for short term, for longer term store at -20°C. Stable for 12 months from date of receipt. |
There has been much recent interest in gene editing and other genetic manipulations by CRISPR-Cas family enzymes. Recent ecosystem and microbiome DNA sequencing and assembly characterized genomes of "huge phages" or megaphages, see Al-Shayeb et al. Nature 578:425-431 (2020) and showed that they contained novel Cas family enzymes. The lab of Nobel prize winner Jennifer Doudna characterized these Cas enzymes and showed that they could be used to perform CRISPR, but had the significant advantage that they were much smaller in molecular size than other Cas family enzymes such as the widely used Cas9 enzymes from S. pyogenes and S. aureus, see Pausch, P. et al. Science 369:333-337 (2020). One of these enzymes is referred to as CasΦ and also known as Cas-12j and was the focus of the Pausch et al. paper. The use of such smaller enzymes leaves more room for other nucleic acid sequences in AAV and other viral vectors which typically have a limited DNA capacity, thus allowing more versatility for new CRISPR based manipulations.
This antibody was raised against a recombinant construct of the CasΦ, specifically the variant listed as CasΦ-2 in reference 2. The host organism for this was Biggievirus Mos11. The 3D structure of this protein is known from cryo-EM studies see Chain A Cas-Phi2. This link also includes the amino acid sequence. The construct was inserted into the eukaryotic expression vector pET30a(+) which adds an N terminal His-tag and some other sequence, underlined below. This sequence includes a thrombin cleavage site (blue), an S-tag affinity peptide (red) and an enterokinase cleavage site (green).
MHHHHHHSSG LVPRGSGMKE TAAAKFERQH MDSPDLGTDD DDKAMADIGS EFMPKPAVES 60
EFSKVLKKHF PGERFRSSYM KRGGKILAAQ GEEAVVAYLQ GKSEEEPPNF QPPAKCHVVT 120
KSRDFAEWPI MKASEAIQRY IYALSTTERA ACKPGKSSES HAAWFAATGV SNHGYSHVQG 180
LNLIFDHTLG RYDGVLKKVQ LRNEKARARL ESINASRADE GLPEIKAEEE EVATNETGHL 240
LQPPGINPSF YVYQTISPQA YRPRDEIVLP PEYAGYVRDP NAPIPLGVVR NRCDIQKGCP 300
GYIPEWQREA GTAISPKTGK AVTVPGLSPK KNKRMRRYWR SEKEKAQDAL LVTVRIGTDW 360
VVIDVRGLLR NARWRTIAPK DISLNALLDL FTGDPVIDVR RNIVTFTYTL DACGTYARKW 420
TLKGKQTKAT LDKLTATQTV ALVAIDLGQT NPISAGISRV TQENGALQCE PLDRFTLPDD 480
LLKDISAYRI AWDRNEEELR ARSVEALPEA QQAEVRALDG VSKETARTQL CADFGLDPKR 540
LPWDKMSSNT TFISEALLSN SVSRDQVFFT PAPKKGAKKK APVEVMRKDR TWARAYKPRL 600
SVEAQKLKNE ALWALKRTSP EYLKLSRRKE ELCRRSINYV IEKTRRRTQC QIVIPVIEDL 660
NVRFFHGSGK RLPGWDNFFT AKKENRWFIQ GLHKAFSDLR THRSFYVFEV RPERTSITCP 720
KCGHCEVGNR DGEAFQCLSC GKTCNADLDV ATHNLTQVAL TGKTMPKREE PRDAQGTAPA 780
RKTKKASKSK APPAEREDQT PAQEPSQTS 809
Molecular weight: 90871.38
Theoretical pI: 9.36
Amino acid composition:
Ala (A) 77 9.5%
Arg (R) 65 8.0%
Asn (N) 26 3.2%
Asp (D) 43 5.3%
Cys (C) 15 1.9%
Gln (Q) 32 4.0%
Glu (E) 58 7.2%
Gly (G) 47 5.8%
His (H) 19 2.3%
Ile (I) 34 4.2%
Leu (L) 60 7.4%
Lys (K) 63 7.8%
Met (M) 11 1.4%
Phe (F) 26 3.2%
Pro (P) 51 6.3%
Ser (S) 50 6.2%
Thr (T) 52 6.4%
Trp (W) 13 1.6%
Tyr (Y) 20 2.5%
Val (V) 47 5.8%
Total number of negatively charged residues (Asp + Glu): 101
Total number of positively charged residues (Arg + Lys): 128
Extinction coefficients in units of M-1 cm-1, at 280 nm measured in water.
Ext. coefficient 102175
Abs 0.1% (=1 g/l) 1.124, assuming all pairs of Cys residues form cystines
Ext. coefficient 101300
Abs 0.1% (=1 g/l) 1.115, assuming all Cys residues are reduced
1. Al-Shayeb, B et al. Clades of huge phages from across Earth’s ecosystems. Nature DOI: 10.1038/s41586-020-2007-4 578:425-531 (2020).
2. Pausch, P. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science DOI: 10.1126/science.abb1400 369:333-337 (2020).
Add a short description for this tabbed section